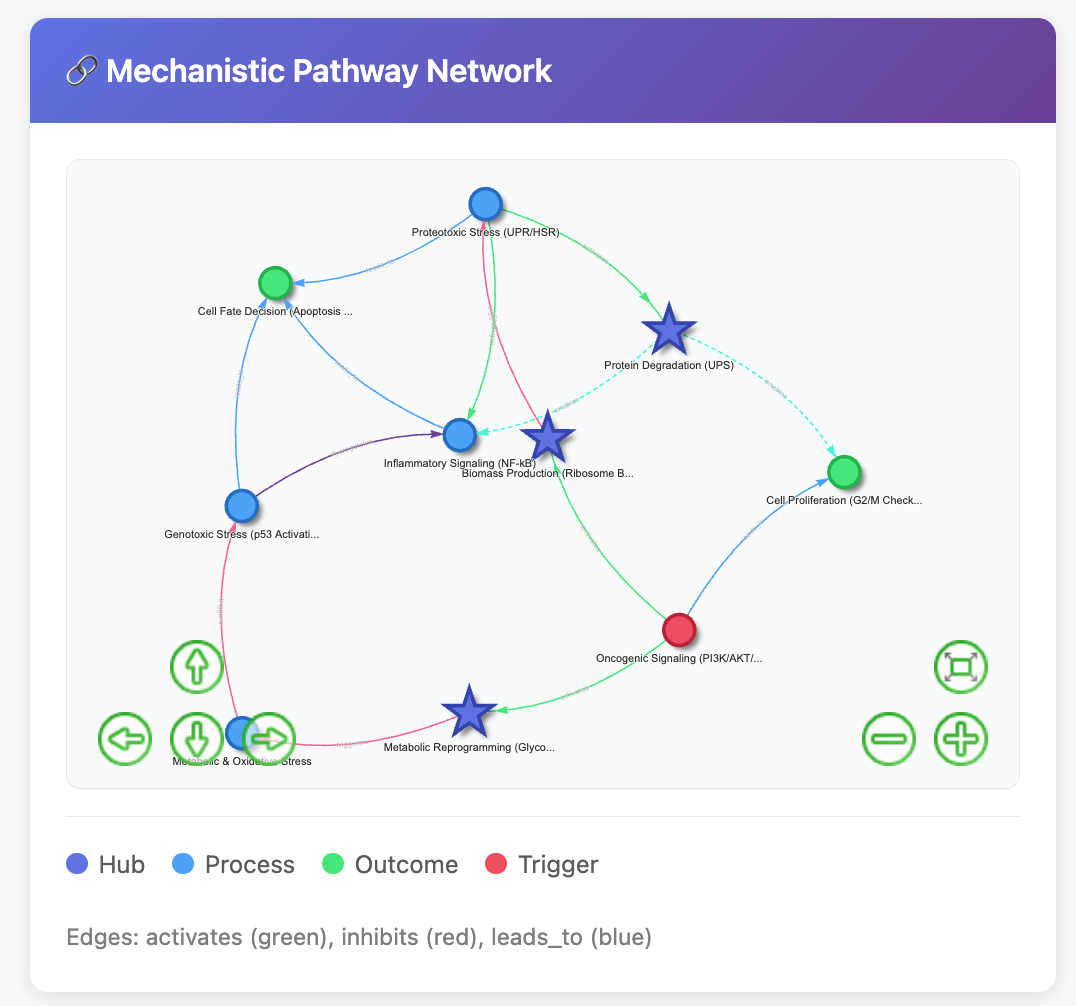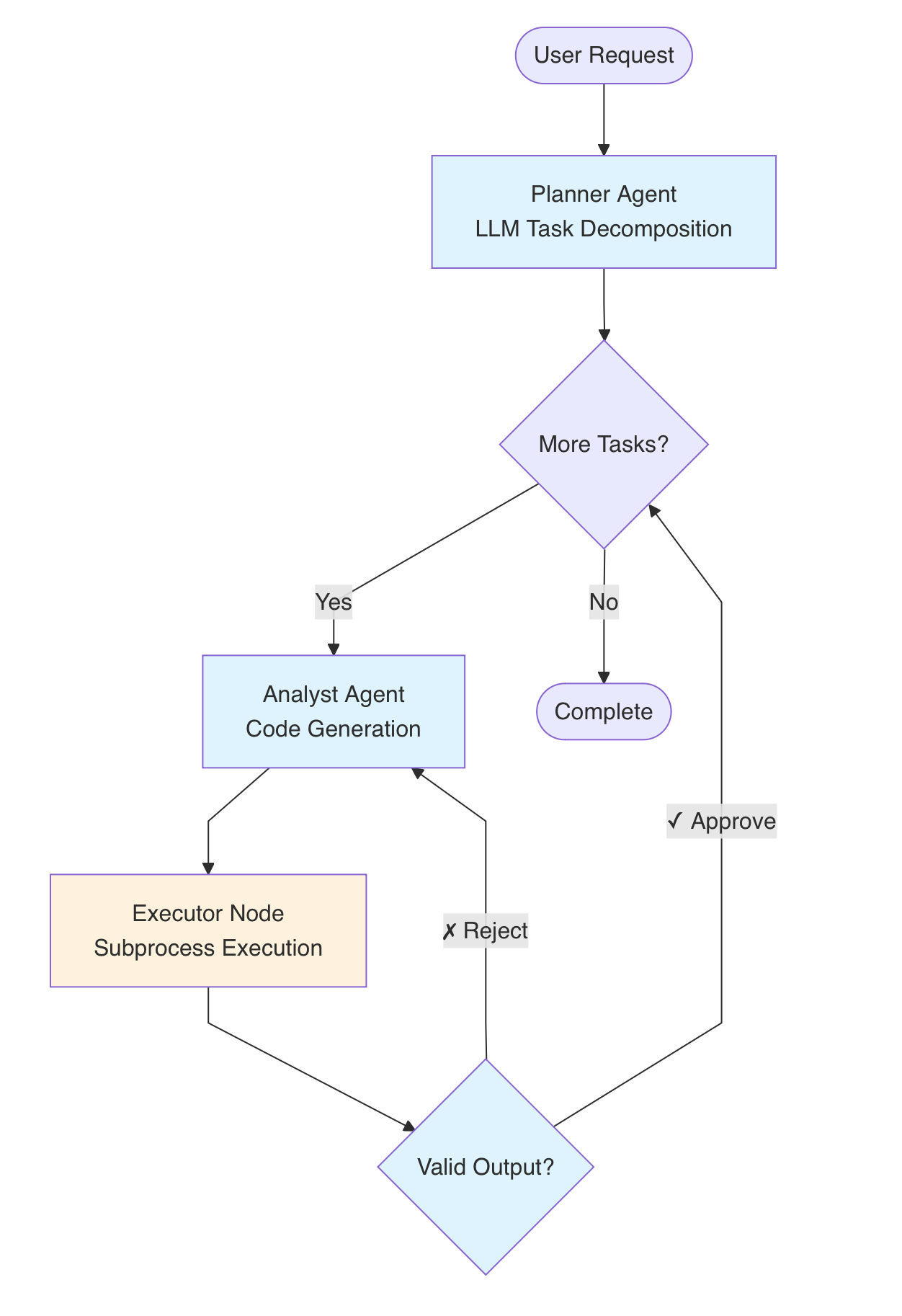
Can AI Match Human Experts at Cell Type Annotation?
Single-cell RNA sequencing has revolutionized how we study biology. A single experiment can profile hundreds of thousands of individual cells, revealing the diversity of cell types in any tissue. But there’s a bottleneck: someone has to make sense of all that data. The standard workflow—quality control, filtering, normalization, clustering, and cell type annotation—requires PhD-level expertise and weeks of hands-on work. Data scientists at research institutions are perpetually backlogged. Scientists wait weeks just to see their first annotated UMAP. ...