You’ve run your experiment. Differential expression identified 500 genes. Gene Set Enrichment Analysis returned 200 significant pathways across three databases. Now what?
This is where most analyses stall. Pathway names are redundant (“cell cycle,” “G2/M checkpoint,” “mitotic spindle” all capture similar biology). Literature context requires hours of PubMed searches. Connecting pathways into mechanistic narratives requires deep domain expertise. And the sheer cognitive load means interpretation is often superficial, biased toward pathways the researcher already knows.
I built a multi-agent system to solve this.
Four Agents, One Biological Story
The system uses four specialized AI agents that work sequentially:
- Context Agent: Clusters redundant pathways using Jaccard similarity on gene overlap, identifying meta-pathways that capture distinct biological processes
- Discovery Agent: Mines literature for each meta-pathway, identifying established mechanisms and novel genes
- Validation Agent: Cross-references findings against experimental data, flagging discrepancies
- Synthesis Agent: Generates mechanistic narratives connecting pathways into testable hypotheses
 Output showing clustered pathways, meta-pathway names, descriptions, established genes, novel genes, and proposed mechanisms. What would take hours of manual curation is generated in under a minute.
Output showing clustered pathways, meta-pathway names, descriptions, established genes, novel genes, and proposed mechanisms. What would take hours of manual curation is generated in under a minute.
Mechanistic Reasoning
The real power emerges when the system connects pathways that humans might not link.
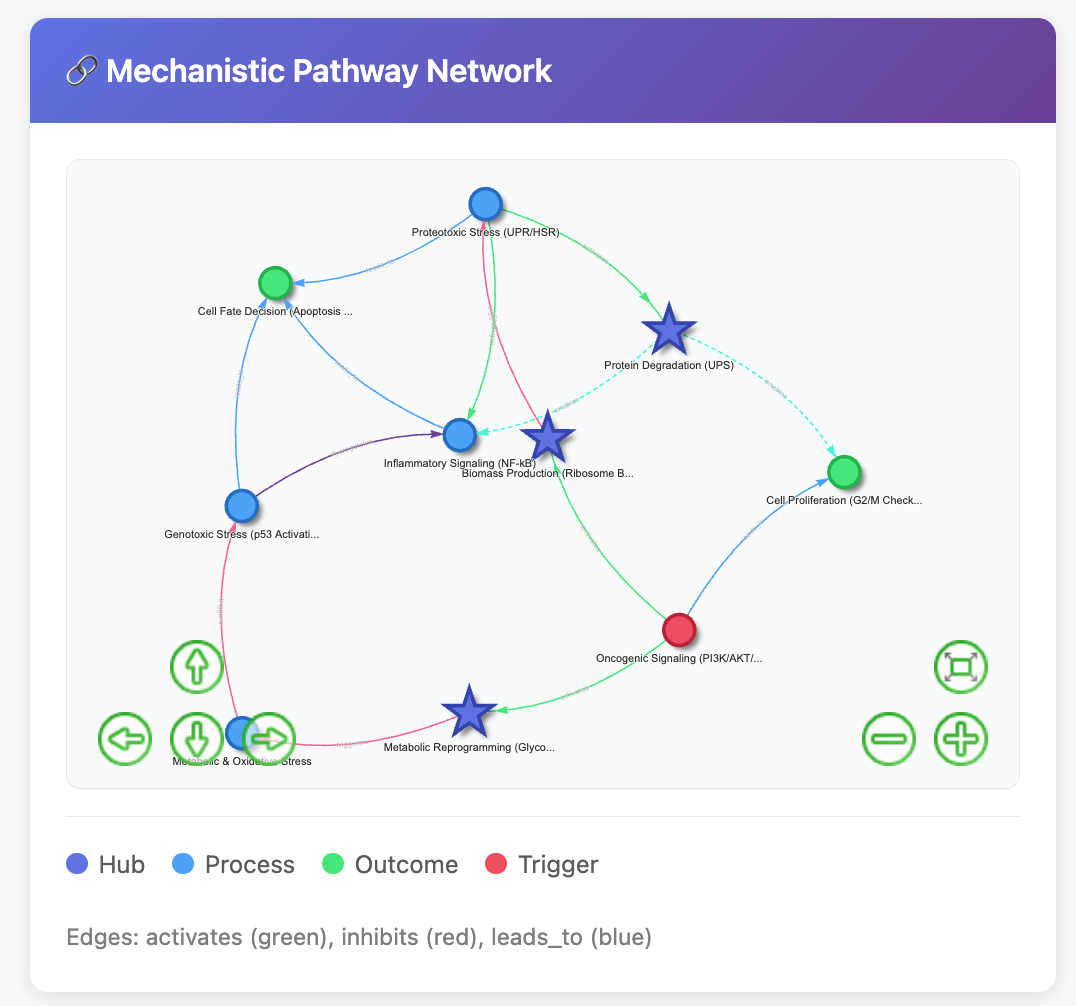 The system generates mechanistic pathway networks showing relationships between processes. Hub nodes (dark blue) represent central regulatory mechanisms. Edges indicate activation (green), inhibition (red), or causal relationships (blue). This visualization emerged from a fibroblast inflammation dataset.
The system generates mechanistic pathway networks showing relationships between processes. Hub nodes (dark blue) represent central regulatory mechanisms. Edges indicate activation (green), inhibition (red), or causal relationships (blue). This visualization emerged from a fibroblast inflammation dataset.
From this network, the system identified NNMT—an enzyme linking metabolism to epigenetics—as a potential driver of fibroblast inflammation. This wasn’t in our hypothesis set going in. The connection emerged from the AI synthesizing pathway co-enrichment patterns with literature context.
Generating Testable Hypotheses
The system doesn’t just summarize—it proposes experiments.
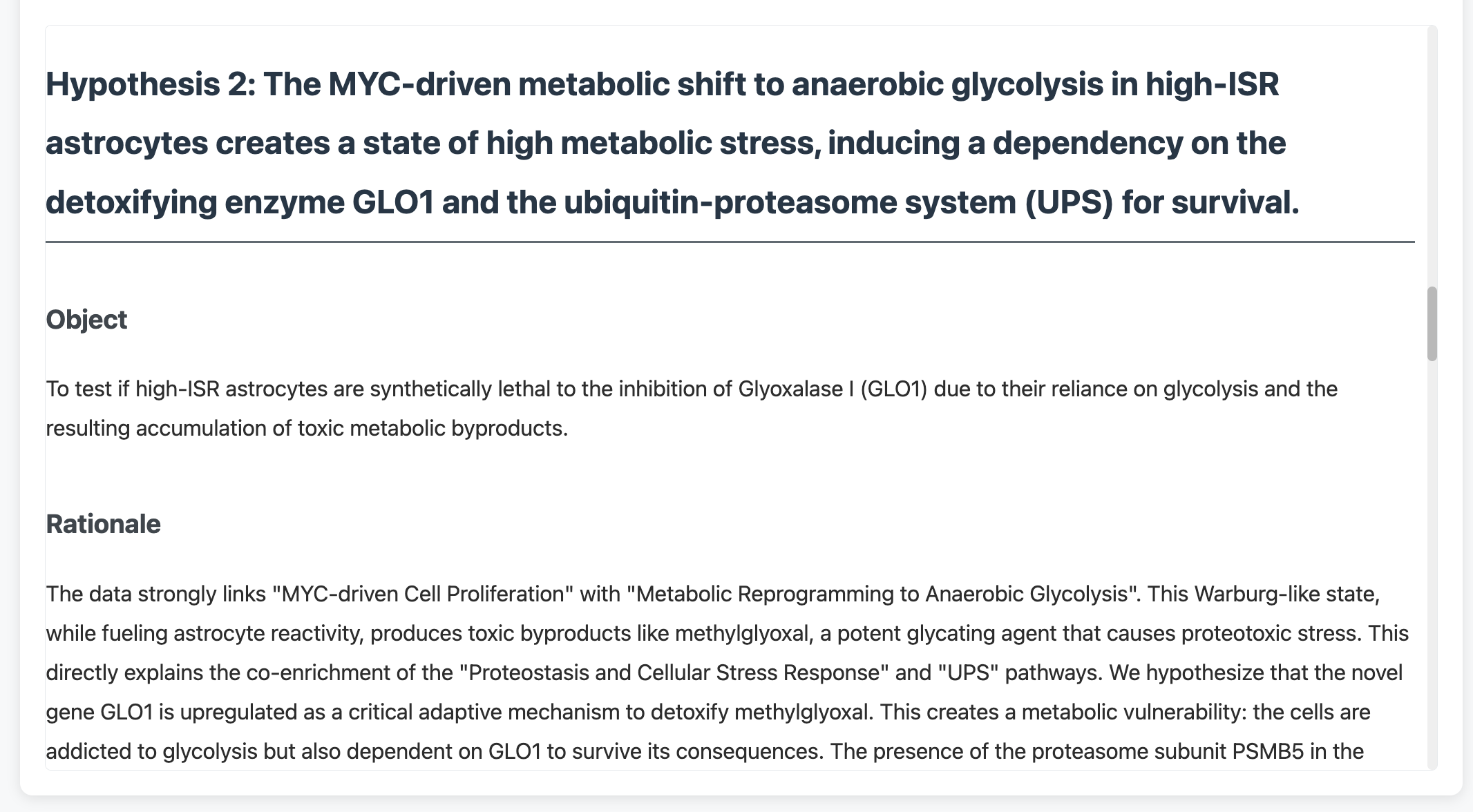 Example hypothesis generated by the system, complete with mechanistic rationale and experimental predictions. These aren’t generic suggestions—they emerge from the specific pathway patterns in your data.
Example hypothesis generated by the system, complete with mechanistic rationale and experimental predictions. These aren’t generic suggestions—they emerge from the specific pathway patterns in your data.
Adoption and Impact
Within the first month, 15 scientists adopted this tool for their analyses. It’s now been used on over 100 reports spanning RNA-seq, proteomics, metabolomics, and spatial transcriptomics.
The key design principle: AI doesn’t replace established bioinformatics methods like GSEA. Instead, it handles the cognitive bottleneck—synthesizing, contextualizing, and connecting—where human bandwidth is the limiting factor.
What takes scientists hours of literature review and pathway curation now happens in under a minute. More importantly, the AI surfaces connections that confirmation bias might cause humans to miss.
